Magnetofection
PolyMag Neo Transfection Reagent
For all nucleic acids transfection
Brand: OZ Biosciences
Available
Free delivery within Singapore only

 SingaporeSG
SingaporeSG ChinaCN
ChinaCN MalaysiaMY
MalaysiaMY IndonesiaID
IndonesiaID MyanmarMM
MyanmarMM_pddqnqv8.png)


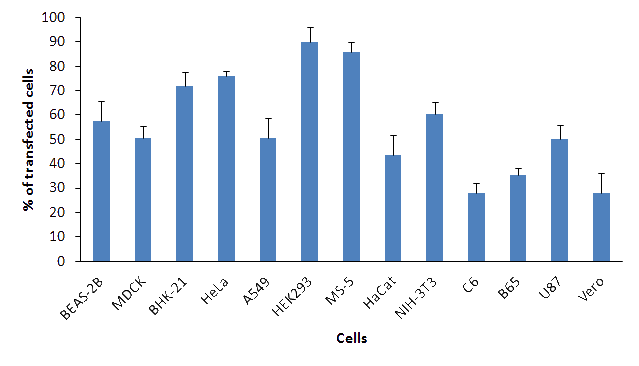 Figure 1: PolyMag Neo transfection efficiency in various cell lines. 1x105 cells were transfected with 0.5 μg / well of pEGFP plasmid DNA in 24-well plates. Transfections were performed with 0.5 μL / well of polyMag Neo reagent. Percentage of transfected cells was measured 24h post transfection by flow cytometry.
Figure 1: PolyMag Neo transfection efficiency in various cell lines. 1x105 cells were transfected with 0.5 μg / well of pEGFP plasmid DNA in 24-well plates. Transfections were performed with 0.5 μL / well of polyMag Neo reagent. Percentage of transfected cells was measured 24h post transfection by flow cytometry.
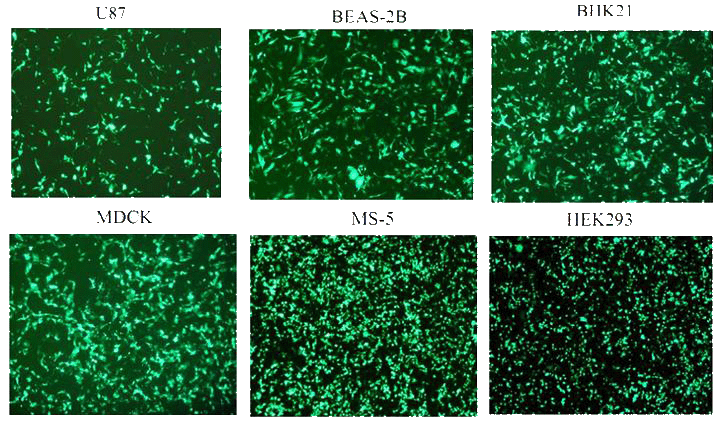 Figure 2: PolyMag Neo transfection efficiency in various cell lines.1x105 cells were transfected with 0.5 μg / well of pEGFP plasmid and 0.5 μL of PolyMag Neo reagent in 24-well plates. EGFP expression was monitored 24h after transfection by fluorescence microscopy.
Figure 2: PolyMag Neo transfection efficiency in various cell lines.1x105 cells were transfected with 0.5 μg / well of pEGFP plasmid and 0.5 μL of PolyMag Neo reagent in 24-well plates. EGFP expression was monitored 24h after transfection by fluorescence microscopy.
_1nglpkag.jpg)
_4oo29y7l.jpg)
_6kqipu0c.jpg)
_uud301p2.jpg)



